Dynamic [15O]H2O-PET: regional analysis of brain, skeletal muscle, and tumour blood flow
Perfusion model
The methods to measure perfusion with [15O]H2O are
based on the principle of exchange of inert gas between blood and tissues
(Kety 1945).
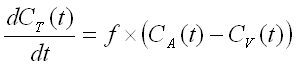
A two compartment (i.e. one tissue compartment) model is used to
describe the kinetics of [15O]H2O (diffusible tracer)
concentration in the tissue, Ct(t), depending on the concentration
in arterial blood, Ca(t), perfusion or blood flow (f), and
the partition coefficient of water, p.
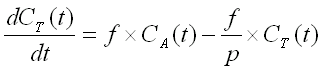
When radioactivity in the volume of interest is measured with PET
(CPET(t)), the
radioactivity of vascular blood inside the
measured volume should also be taken into account;
Va is the arterial volume fraction in tissue:
![]()
Model fit and simulation programs
Perfusion model parameters can be estimated (fitted) using fit_h2o. This program can estimate four parameters: blood flow, partition coefficient, arterial volume fraction, and the time delay. Any of the parameters can be fixed to a certain value by editing the program parameter file. Regional tissue TACs can be simulated using program b2t_h2o.
Steps of perfusion calculation:
The following steps can be done in MS Windows command prompt window, or Solaris terminal window on either SUN or PC platform.
1. Pre-processing of arterial blood curve
Arterial blood data from the on-line sampler needs to be processed before it can be used as input function in the perfusion calculation.
If you are working in TPC, use the script water_input to process the on-line detector (blood pump) data prior to the analysis. It requires the countrate curve (*.cr) or similar data for the time delay correction; please note that in some organs you should not use countrate curve, but a TAC from region-of-interest instead.
In some occations you can create arterial blood curve from the dynamic PET image. In this case no corrections need to be done.
2. Preparation of regional tissue TAC data
This is explained in detail elsewhere. In short: draw ROIs and calculate regional TACs from dynamic images, and calculate averages over planes and regions if required. If you have regional TACs in old format (*.roi.kbq), convert those to DFT format using program nci2dft.
Before proceeding, make sure that both the blood and tissue data are in the
same calibration units (preferably kBq/ml)
and that the time unit is sec.
Image data from HR+ and PET-CT may originally be in units Bq/ml.
You can view the files in text editor, or
see and
convert the units with dftunit.
3. Adding weights to regional tissue TAC data
Weights should be added to tissue data file using dftweigh. Weights can be calculated based on either SIF file (*dy1.img.sif) or the average tissue curves. The weights are not absolute, but only relational to each time frame in the TAC.
If weights are extracted from the SIF file, the command could be e.g.:
dftweigh uo0268.dft uo0268dy1.img.sif O-15
If SIF file is not used, the command would be:
dftweigh uo0268.dft
4. Calculation of blood flow
The fit_h2o program can be run with following command line parameters:
- corrected blood TAC file
- regional tissue TAC file
- length of study to be used in the fitting (usually a large number to allow fitting to the end of data)
- name for result file; note that an existing result file is overwritten
In addition, the fitted TACs can be saved for later use by specifying a filename for it.
For example, the command could be like this:
fit_h2o uo0268ab.kbq uo0268.dft 999 uo0268.res uo0268fit.dat
The result file can be saved in HTML format, if the extension is set to .html.
If you need to fix a parameter or set lower and upper limits for it, those can be specified in a parameter file that is given with option -i. If you
do not have the parameter file, you can let the program make it for you by
entering on the command line only the program name, and the name of the newparameter file, e.g.
fit_h2o -i=owater.set
Then you can edit the file with any text editor, e.g. Notepad (if you use Wordpad or Word etc, be careful not to save any formatting codes). If you want to fix one or more of the parameters, e.g. partition coefficient to, say, p=0.8, or have already fitted time delay and thus delayT=0, you should set both limits to this same value. You should use the same parameter settings file for your whole study material.
